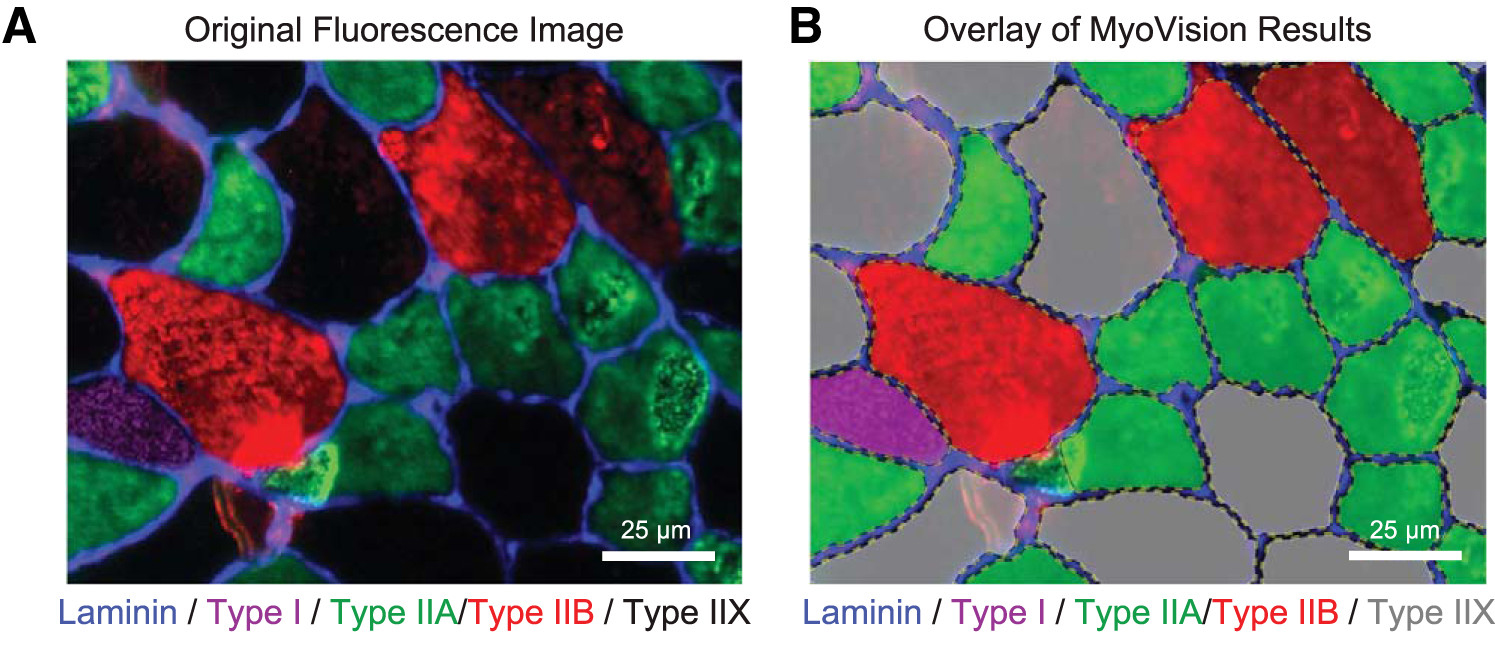
Analysis of skeletal muscle cross sections is an important experimental technique in muscle biology. Many aspects of immunohistochemistry and fluorescence microscopy can now be automated, but most image quantification techniques still require extensive human input, slowing progress and introducing the possibility of user bias. MyoVision is a new software package that was developed to overcome these limitations. The software improves upon previously reported automatic techniques and analyzes images without requiring significant human input and correction. When compared with data derived by manual quantification, MyoVision achieves an accuracy of ≥94% for basic measurements such as fiber number, fiber type distribution, fiber cross-sectional area, and myonuclear number. Scientists can download the software free from www.MyoVision.org and use it to automate the analysis of their own experimental data. This will improve the efficiency and consistency of the analysis of muscle cross sections and help to reduce the burden of routine image quantification in muscle biology. NEW & NOTEWORTHY Scientists currently analyze images of immunofluorescently labeled skeletal muscle using time-consuming techniques that require sustained human supervision. As well as being inefficient, these techniques can increase variability in studies that quantify morphological adaptations of skeletal muscle at the cellular level. MyoVision is new software that overcomes these limitations by performing high-content analysis of muscle cross sections with minimal manual input. It is open source and freely available.